1. What is CellREADR
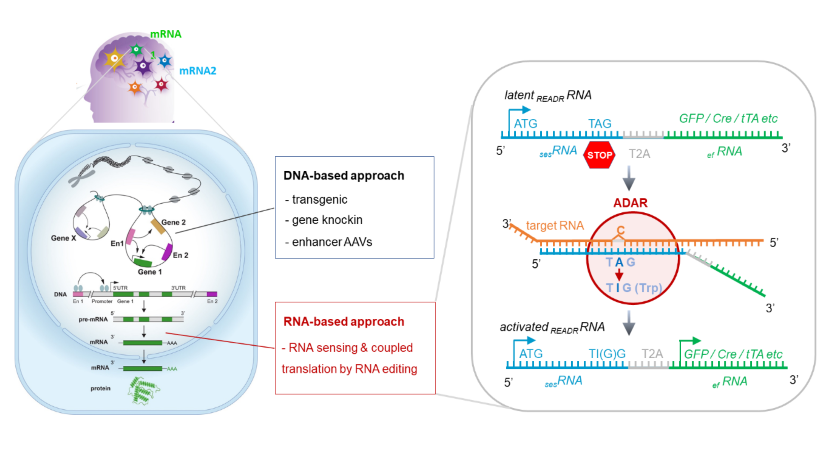
CellREADR stands for Cell access through RNA sensing by Endogenous ADAR (adenosine deaminase acting on RNA), and represents a new generation of RNA-based cell monitoring and manipulation technology. CellREADR is implemented as a modular RNA molecule that can detect the presence of a cellular RNA and then triggers the translation of a desired effector protein to monitor and manipulate the cell; the coupling of RNA sensing and effector translation is mediated by RNA editing, catalyzed by a cell endogenous enzyme (ADAR) ubiquitous to all metazoan cells.
The diversity of cells underlies the diversity of life forms and of physiological systems within individual organisms. RNAs are the universal mediator of genetic information underlying cell diversity to shape tissue organization and function across species and life spans. High-throughput single-cell RNA sequencing promise to identify all molecularly-defined cell types in human and other organisms. Beyond transcriptome profiling, it is necessary to monitor and manipulate each and every cell type to identify their specific roles in tissue architecture and system function. Cell-specific monitoring and intervention is also key to precision diagnosis and treatment of human diseases. To achieve these, we need cell type technologies that are specific, facile, scalable, and generalizable across species.
Most current genetic approaches to accessing cell types rely on DNA-based transcriptional regulatory elements to mimic cell-specific RNA expression either through germline engineering or viral vectors driven by transcriptional enhancer elements. However, germline engineering is cumbersome and difficult to scale and generalize; enhancers are challenging to identify and validate due to their complex relationship to cell types. Ultimately, all DNA-based approaches are inherently indirect in attempting to recapitulate cell-specific RNA expression patterns. CellREADR bypasses the entire DNA-based transcriptional process and harnesses an RNA sensing and editing mechanism, which directly detects cell-specific RNAs and then switches on translation of effector proteins to monitor and control the cell. Deployed as a single RNA molecule operating through Watson-Crick base pairing, CellREADR as a cell engineering technology is inherently specific, easy, scalable, programmable, and generalizable across species.
2. Properties
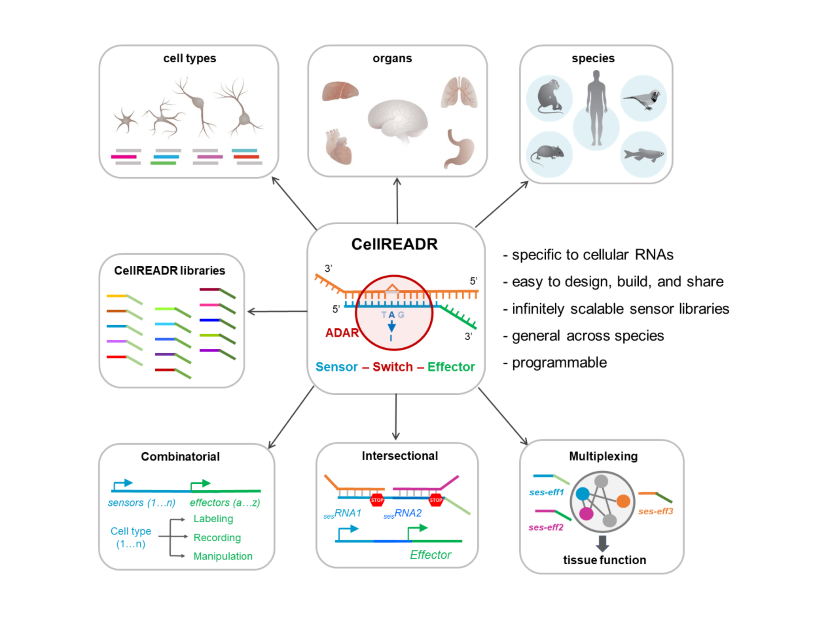
As CellREADR operates through RNA sequence base-pairing and RNA editing mediated by cell endogenous ADAR, it has the following properties:
1) specific to cells defined by RNA expression;
2) easy to build, use, and disseminate;
3) scalable for targeting many RNAs and RNA-defined cells in many tissues;
4) generalizable to most animal species including human;
5) sequence-programmable for combinatorial, intersectional, and multiplexed cell targeting and control
Figure 2. CellREADR properties
3. Delivery
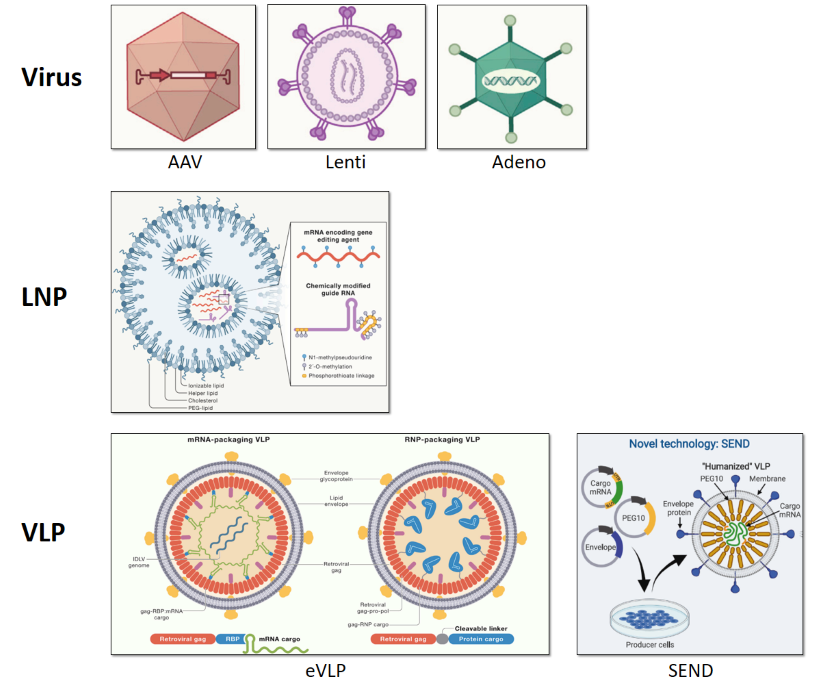
CellREADR can be delivered for in vivo application either as DNA expression vectors or as RNA directly.
Delivery methods include:
- viral vectors: adeno-associated virus (AAVs), adenovirus, lenti-virus etc
- lipid nanoparticles (LNPs)
- viral-like particles (VLPs)
Here is a review on therapeutic in vivo deliver system (Raguram et al., Cell 2022).
Figure 3. Emerging delivery methods for CellREADR
4. Applications
As the RNA sensing domain and payload translation domain can be independently and coordinately programmed, CellREADR has exceptionally wide applications in biomedical research and medicine. These include but are not limited to:
- visualize and monitor cell types or cell states, e.g. by expressing fluorescent proteins or physiological reporter proteins;
- modulate the physiology and function of cell types, e.g. by expressing ion channels, signaling proteins;
- reprogram a cell identity, e.g. by expressing master transcription factors;
- modulate cell-cell interactions, e.g. by expressing cell adhesion molecules or intercellular signaling proteins;
- eliminate deleterious cells (cancer cells or viral infected cells), e.g. by expressing apoptotic protein or receptors for immune killer cells
5. Reagents and Resources
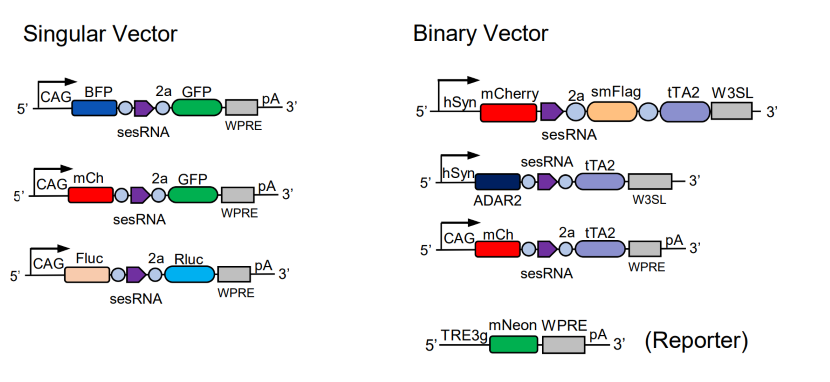
We designed both a singular and a binary vector system for CellREADR.
The singular vector directly couples RNA detection with tool protein translation.
In the binary system, the Readr vector couples RNA detection to the translation of a transcriptional activator tTA, and the Reporter vector contains a tTA-driven promoter (TRE3g) to amplify payload expression.
Thus the binary vectors provide amplification as well as combinatorial flexibility for expressing different tool genes by pairing Readr and Reporter vectors. The relevant plasmids are available from Addgene.
Figure 4. Vectors for CellREADR
6. How to Design a sesRNA
sesRNA (sense-edit-switch RNA, i.e. RNA sensor) is the key determinant of the specificity and efficiency of CellREADR. It is designed to detect the presence of a specific cellular RNA through base pairing and recruit ADAR-mediated RNA editing to trigger the translation of an effector protein. We have yet to identify sesRNA sequence features that facilitate the specificity and efficiency of RNA sensing and editing. Currently we often test several sesRNAs for a target RNA to identify ones with sufficient specificity and efficiency. Based on empirical data, here we describe our current procedure for sesRNA design. We also provide an online-tool for designing sesRNAs for human and mice RNAs.
1) sesRNA is complementary, i.e. anti-sense, to a cellular coding or non-coding RNA sequence;
2) sesRNAs can be directed to any region of a cellular transcript, including exons, introns, 5’ and 3’untranslated regions (UTRs), or mature mRNA after splicing;
3) sesRNA is followed by an efRNA arranged in the same translation reading frame;
4) The length of a sesRNA is typically 200-300 nucleotides;
5) One or more STOP codons (TAG) are placed near the center of the sesRNA (between ~80-220 nucleotides from 5’ end);
6) find a 5’-CCA-3’ sequence in the target RNA as seed to design a complementary sesRNA centered around the 5’-TGG-3’ sequence; then replace 5’-TGG-3’ in the sesRNA sequence with 5’-TAG-3’, so that a mismatch A-C will be introduced when sesRNA base pairs with the target RNA;
7) to ensure that there are no other STOP codons in the sesRNA, all other TAG, TAA, TGA sequences in sesRNA are converted to TAC, TAC, TCA, respectively; preferably the converted STOP codons are not less than 10 nucleotides from the TAG designed in step 5;
8) there should be no in-frame ATG (translation initiation codon) after TAG defined in step 5 to exclude the possibility of unintended translation initiation;
9) avoid sesRNA with complex secondary structures.
10) avoid designing sesRNA with sequence homologous to other unintended RNAs or genes
Future efforts will combine experimental and computational approaches to screen and validate large sesRNA libraries and to develop algorithms for improved sesRNA design.