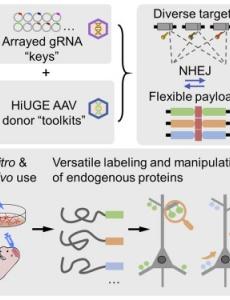
A new Soderling Lab paper reports a simplified approach for flexibly tagging endogenous proteins in vitro & in vivo. They demonstrate that universal CRISPR-Cas9-based homology-independent universal genome engineering (HiUGE) donors enable rapid alterations of proteins in vitro or in vivo for protein labeling and dynamic visualization, neural-circuit-specific protein modification, subcellular rerouting and sequestration, and truncation-based structure-function analysis. Thus, the “plug-and-play” nature of HiUGE enables high-throughput and modular analysis of mechanisms driving protein functions in cellular neurobiology.