Franks Lab
The Problem
One of the key features of brain function is to extract patterns from noisy sensory input, update expectations based on this experience, and then use that knowledge to adaptively guide behavior. We call these processes Perception, Learning, and Memory. In the Franks Lab, we study how these processes are implemented in the brain—and what happens when they break down in disease. Using the mouse olfactory system, we investigate how neural circuits transform raw sensory input into stable odor representations, and how these representations are modified by experience, stored over time, and used to support adaptive behavior.
In fact, deficits in odor identification and recognition are among the earliest and strongest predictors of neurodegenerative diseases like Alzheimer’s and Parkinson’s. By uncovering the basic principles of how the brain learns, stores, and retrieves sensory information, our work provides a foundation for understanding what goes wrong in disease and how we might intervene.
Our Approach
We integrate large-scale electrophysiology, optogenetics, in vitro slice physiology, behavior, and computational modeling to causally link behavior to circuit dynamics to synaptic mechanisms. The olfactory system offers a highly tractable circuit for this approach because we can directly and precisely control the sensory input to cortical circuits. This allows us to characterize how information is transformed within the piriform cortex, and to uncover the neural mechanisms supporting these transformations by manipulating and measuring activity in defined neuron populations.
Our goals are to elucidate the piriform cortex’s role in odor-guided behavior and to identify fundamental principles of cortical computation that apply across sensory modalities and behavioral states—including those disrupted in neurological and neurodegenerative disease.
Innovations and Discoveries
We have extensively characterized the excitatory and inhibitory synaptic organization of the piriform cortex, contributing to a quantitative wiring diagram of this evolutionarily conserved circuit. By combining in vivo recordings, optogenetics, and modeling, we revealed how input from the olfactory bulb is integrated and shaped by local inhibition and recurrent connectivity, providing a mechanistic foundation for how sensory representations are formed and refined in cortex.
We discovered that odor identity and odor intensity are encoded by distinct features of the population response. While odor identity is determined by which neurons are activated, intensity is encoded in the temporal synchrony of the response. Importantly, we showed that piriform cortex circuitry transforms these features into concentration-invariant representations—ensuring that the identity of an odor remains stable despite changes in stimulus strength.
Most recently, we’ve shown that piriform cortex implements a phase-to-rate transformation: converting temporal input from the olfactory bulb, aligned to the respiration cycle, into reliable ensemble codes. This transformation supports rapid and robust odor recognition and may reflect a more general strategy by which cortical circuits stabilize temporally structured input across sensory systems.
Franks Lab Research
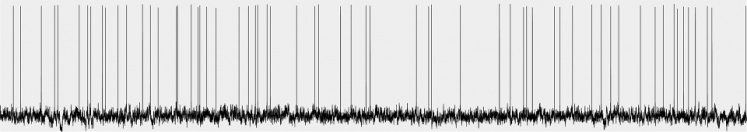
Odors are represented as ensembles of cortical neurons in mouse olfactory, or piriform cortex. These cells receive diffuse and overlapping afferent sensory inputs from the olfactory bulb. They also form a rich, interconnected recurrent network, where a given cortical pyramidal cell receives inputs from hundreds of other pyramidal cells distributed across piriform cortex. Together, the sensory and recurrent inputs generate odor-specific cortical ensembles. We ask: how are these ensembles formed; how does the cortex discriminate between similar odorants to generate wholly different cortical ensembles; how do cortical ensembles represent odorants at different concentrations; and how are novel versus learned odorants represented?
In combination with various optogenetic and molecular tools to perturb defined elements of the cortical circuit, we use in vivo and in vitro electrophysiology and optical imaging of neural activity, together with behavioral and computational analyses, to understand the generation of odor-specific cortical odor ensembles, and the roles of different cell types within the circuit in shaping these.
Franks Lab Publications
Olfactory Circuits
Recurrent circuitry is required to stabilize piriform cortex odor representations across brain states.
Bolding KA, Nagappan S, Han, BX, Wang F & Franks KM. (2020) eLife 2020;9:e53125.
doi: 10.7554/eLife.53125. PMID: 32662420
Structure and flexibility in cortical representations of odour space.
Pashkovski SL, Iurilli G, Brann D, Chicharro D, Drummey K, Franks KM, Panzeri S, Datta SR. (2020)
Nature 583:253-258. doi: 10.1038/s41586-020-2451-1. PMID: 32612230.
Odor coding in piriform cortex: mechanistic insights into distributed coding.
Blazing RM & Franks KM. (2020) Curr. Opin. Neurobiol. 64: 96-102.
doi: 10.1016/j.conb.2020.03.001. PMID: 32422571.
Th17 lymphocytes drive vascular and neuronal deficits in a mouse model of postinfectious autoimmune encephalitis.
Platt MP, Bolding KA, Wayne CR, Chaudry S, Cutforth T, Franks KM & Agalliu D. (2020) PNAS 117: 6708—16.
doi: 10.1073/pnas.1911097117. PMID: 32161123.
Recurrent cortical circuits implement concentration-invariant odor coding.
Bolding KA & Franks KM. (2018) Science 361: eaat6904.
doi: 10.1126/science.aat6904. PMID: 30213885.
A transformation from temporal to ensemble coding in a model of piriform cortex.
Stern M, Bolding KA, Abbott LF & Franks KM. (2018) eLife 10.7554/eLife.34831.
doi: 10.7554/eLife.34831. PMID: 29595470.
Learning: Plasticity without Stabilization in Olfactory Cortex.
Nagappan S & Franks KM. (2018) Curr. Biol. 28: R23-R25.
doi: 10.1016/j.cub.2017.11.010. PMID: 29316416.
Complementary codes for odor identity and odor intensity in olfactory cortex.
Bolding KA & Franks KM. (2017) eLife 10.7554/eLife.22630.
Odor identity coding by distributed ensembles of neurons in the mouse olfactory cortex.
Roland B, Deneux T, Franks KM, Bathellier B, Fleischmann A. (2017) eLife 10.7554/eLife.26337.
A transformation from temporal to ensemble coding in a model of piriform cortex.
Stern M, Bolding KA, Abbott LF & Franks KM. (2017) bioRxiv.
Massive normalization of olfactory bulb output in mice with a “monoclonal nose.”
Roland B, Jordan R, Sosulski DL, Diodato A, Fukunaga I, Wickersham I, Franks KM, Schaefer AT & Fleischmann A. (2016) eLife 10:7554/eLife.16335.
I Want It All and I Want It Now: How a Neural Circuit Encodes Odor with Speed and Accuracy.
Franks KM. (2015) Neuron. 88: 852-4.
Multi-step signaling from sensory neurons onto olfactory bulb mitral cells.
Gire DH*, Franks KM*, Zak JD, Tanaka KF, Whitesell JD, Mulligan AA, Hen R, Schoppa NE. (2012) J. Neurosci. 32: 2964-75.
Recurrent circuitry dynamically shapes the activation of piriform cortex.
Franks KM, Russo MJ, Sosulski DL, Mulligan AA, Siegelbaum SA, Axel, R. (2011) . Neuron. 72: 49-56.
Mice with a "monoclonal nose": perturbations in an olfactory map impair odor discrimination.
Fleischmann A, Shykind BM, Sosulski DL, Franks KM, Glinka ME, Mei DF, Sun Y, Kirkland J, Mendelsohn M, Albers MW, Axel R. (2008) Neuron. 60: 1068-81.
Strong single-fiber sensory inputs to olfactory cortex: implications for olfactory coding.
Franks KM, Isaacson JS. (2006) Neuron. 49: 357-363.
Synapse specific downregulation of NMDA receptors by early experience: a critical period for plasticity of sensory input to olfactory cortex.
Franks KM, Isaacson JS. (2005) Neuron. 47: 101-114.
* denotes equal authorship
Technology Development
Single-cell genetic transfection via in vivo whole-cell recording: bridging physiology, genetics and connectomics.
Rancz EA*, Franks KM*, Schwartz, M Schaefer AT, Seeburg P, Margrie TW. (2011) Nature Neuroscience. 14: 527-532.
Older Publications
NGF is essential for hippocampal plasticity and learning.
Conner JM*, Franks KM*, Titterness AK*, Russell K, Merrill DA, Christie BR, Sejnowski TJ, Tuszynski MH. (2009) J. Neurosci. 29: 10883-10889.
Calmodulin activation by calcium transients in the postsynaptic density of dendritic spines.
Keller DK, Franks KM, Bartol TM, Sejnowski TJ. (2008) PLoS One. 3: e2045.
Independent sources of synaptic variability at single glutamatergic synapses.
Franks KM, Stevens CF, Sejnowski TJ. (2003) J. Neurosci. 23: 3186-3195.
Complexity of calcium signaling in synaptic spines.
Franks KM, Sejnowski TJ. (2002) Bioessays. 24: 1130-1144.
A Monte Carlo model reveals independent signaling at central glutamatergic synapses.
Franks KM*, Bartol TM*, Sejnowski, TJ. (2002) Biophys J. 83: 2333-2348.
An MCell model of calcium dynamics and frequency-dependence of calmodulin activation in dendritic spines.
Franks KM, Bartol TM, Sejnowski TJ. (2001). Neurocomputing. 38-40: 9-16.
Synaptic plasticity in morphologically identified CA1 Stratum Radiatum interneurons and projection cells.
Christie BR, Franks KM, Seamans JK, Saga K, Sejnowski, TJ. (2000). Hippocampus. 10: 678-683.
Ryu, Brendan, Shivathmihai Nagappan, Fernando Santos-Valencia, Psyche Lee, Erica Rodriguez, Meredith Lackie, Jun Takatoh, and Kevin M. Franks. “Chronic loss of inhibition in piriform cortex following brief, daily optogenetic stimulation.” Cell Rep 35, no. 3 (April 20, 2021): 109001. https://doi.org/10.1016/j.celrep.2021.109001.
Russo, Marco J., Kevin M. Franks, Roxanne Oghaz, Richard Axel, and Steven A. Siegelbaum. “Synaptic Organization of Anterior Olfactory Nucleus Inputs to Piriform Cortex.” J Neurosci 40, no. 49 (December 2, 2020): 9414–25. https://doi.org/10.1523/JNEUROSCI.0965-20.2020.
Blazing, Robin M., and Kevin M. Franks. “Neuroscience: Illuminating Principles of Odor Coding.” Curr Biol 30, no. 20 (October 19, 2020): R1279–81. https://doi.org/10.1016/j.cub.2020.08.024.
Blazing, Robin M., and Kevin M. Franks. “Odor coding in piriform cortex: mechanistic insights into distributed coding.” Curr Opin Neurobiol 64 (October 2020): 96–102. https://doi.org/10.1016/j.conb.2020.03.001.
Pashkovski, Stan L., Giuliano Iurilli, David Brann, Daniel Chicharro, Kristen Drummey, Kevin M. Franks, Stefano Panzeri, and Sandeep Robert Datta. “Author Correction: Structure and flexibility in cortical representations of odour space.” Nature 584, no. 7822 (August 2020): E38. https://doi.org/10.1038/s41586-020-2615-z.
Bolding, Kevin A., Shivathmihai Nagappan, Bao-Xia Han, Fan Wang, and Kevin M. Franks. “Recurrent circuitry is required to stabilize piriform cortex odor representations across brain states.” Elife 9 (July 14, 2020). https://doi.org/10.7554/eLife.53125.
Pashkovski, Stan L., Giuliano Iurilli, David Brann, Daniel Chicharro, Kristen Drummey, Kevin M. Franks, Stefano Panzeri, and Sandeep Robert Datta. “Structure and flexibility in cortical representations of odour space.” Nature 583, no. 7815 (July 2020): 253–58. https://doi.org/10.1038/s41586-020-2451-1.
Platt, Maryann P., Kevin A. Bolding, Charlotte R. Wayne, Sarah Chaudhry, Tyler Cutforth, Kevin M. Franks, and Dritan Agalliu. “Th17 lymphocytes drive vascular and neuronal deficits in a mouse model of postinfectious autoimmune encephalitis.” Proc Natl Acad Sci U S A 117, no. 12 (March 24, 2020): 6708–16. https://doi.org/10.1073/pnas.1911097117.
Bolding, Kevin A., and Kevin M. Franks. “Recurrent cortical circuits implement concentration-invariant odor coding.” Science 361, no. 6407 (September 14, 2018). https://doi.org/10.1126/science.aat6904.
Stern, Merav, Kevin A. Bolding, L. F. Abbott, and Kevin M. Franks. “A transformation from temporal to ensemble coding in a model of piriform cortex.” Elife 7 (March 29, 2018). https://doi.org/10.7554/eLife.34831.
Franks Lab Members
Lab Alumni
Postdocs:
Kevin Bolding (2013-2019; Research Associate, Davison Lab, Boston University)
Erica Rodriguez (2018; Postdoc, Salzman lab, Columbia University)
Graduate Students:
(none yet)
Honors Students:
Brendan Ryu (with distinction)
Visiting Scholars:
Maryanne Platt (visiting graduate student, Columbia University)
Benjamin Roland (visiting graduate student, Collège de France)
Technicians:
Samuel Montgomery (traveling)
Brendan Ryu (Medical student, Hofstra University)
Mandy Munsch (Bioengineering Ph.D. program, UNC)
Meredith Lackie (Medical student, University of Vermont)
Melissa Franch (Ph.D. program, UT Health Houston)
Mark Dalgetty (Medical student, University of South Carolina)
Psyche Lee (retired)
Undergraduates:
Kanav Chhabra (Venture For America)
Keara Darrah (Medical student, UCLA)
Hannah McMullan (Pharmacology Ph.D. program at U. Minnesota)
Eric Song (Software Engineer, Amazon)
Sade Abiodun (Neuroscience Ph.D. program at Princeton U.)
Surabhi Beriwal (Rhodes iiD, Duke)
Katherine Kim (Duke undergraduate)
Franks Lab Positions
Join Franks Lab
- Are you talented, creative and highly motivated?
We are always looking for the right people to join our team.
- Are you eager to understand how neural circuits in the brain are wired, and what computational power these wiring patterns confer upon them?
We are using a variety of techniques to probe the logic of cortical odor representations, including in vivo and in vitro electrophysiology and optical imaging, optogenetics, viral and genetic disruption of cortical circuits, and behavioral analyses.
- Do you want to work in a focused lab that is also part of a greater neuroscience community?
We are part of a growing department with a highly interactive core group of labs all studying neural circuits in different systems and brain areas.
If you think that this might be just the right scientific environment for you, please send a C.V., a brief description of your past work and future research interests, and the names of 3 references to franks@neuro.duke.edu.



