Meyer Lab
Education
Ph.D., Northwestern University, 2010
The Meyer Lab studies RNA regulatory pathways in the nervous system. We have a particular interest in understanding how RNA methylation (m6A) controls gene expression programs in the brain to contribute to human health and disease.
 The chemical building blocks that make up RNA are central to its function. For a long time, it was thought that RNAs were made up primarily of four main bases-- A, G, C, and U. Recently, however, we and others have shown that modified forms of these bases also exist and are found in thousands of cellular mRNAs.
The chemical building blocks that make up RNA are central to its function. For a long time, it was thought that RNAs were made up primarily of four main bases-- A, G, C, and U. Recently, however, we and others have shown that modified forms of these bases also exist and are found in thousands of cellular mRNAs.
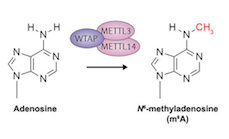 One modified base which we have found to be particularly prevalent throughout the transcriptome is N6-methyladenosine, or m6A, which occurs when A residues become methylated.
One modified base which we have found to be particularly prevalent throughout the transcriptome is N6-methyladenosine, or m6A, which occurs when A residues become methylated.
Many studies in our laboratory focus on m6A and how it contributes to fundamental aspects of gene expression control. We are also interested in understanding how m6A as well as other RNA modifications shape gene expression programs in the nervous system.
Lab Members
Meyer Publications
Meyer, K.D., Patil, D.P., Zhou, J., Zinoviev, A., Skabkin, M.A., Elemento, O., Pestova, T.V., Qian, S., Jaffrey, S.R. 5'UTR m6A promotes cap-independent translation. Cell 163 (4): 999-1010, 2015. (Previewed by Sarah Mitchell and Roy Parker: Cell).
Satterlee, J.S., Basanta-Sanchez, M., Blanco, S., Li, J.B., Meyer, K.D., Pollock, J., Sadri-Vakili, G., Rybak-Wolf, A. Novel RNA modifications in the nervous system: form and function. J Neurosci 34 (46): 15170-15177, 2014.
Meyer, K.D. and Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nature Reviews Molecular Cell Biology 15 (5): 313-326, 2014.
Hess, M.E., Hess, S., Meyer, K.D., Verhagen, L.A., Koch, L., Brönneke, H.S., Dietrich, M.O., Jordan, S.D., Saletore, Y., Elemento, O., Belgardt, B.F., Franz, T., Horvath, T.L., Rüther U., Jaffrey, S.R., Kloppenburg, P., Brüning, J.C. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nature Neuroscience 16 (8): 1042-1048, 2013.
Saletore, Y., Meyer, K.D., Korlach, J., Vilfan, I.D., Jaffrey, S.R., Mason, C.E. The birth of the epitranscriptome: deciphering the function of RNA modifications. Genome Biology 13 (10): 175, 2012.
Meyer, K.D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C.E., Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals pervasive adenosine methylation in 3’ UTRs. Cell 149 (7): 1635-1646, 2012.

