Yang Lab
Drosophila Learning
We recently started to explore the genetic and circuit basis of operant learning in Drosophila. In particular, we are focusing on uncovering genes and circuit mechanisms that lead to superior capacity to learn, using a new optogenetics-assisted approach we have developed. Three important developments laid the foundation for this approach:
First, we believe to have discovered the Drosophila equivalent of the mammalian medial forebrain bundle (MFB). Using a GAL4-based screen, we found a group of neurons whose optogenetic activation is profoundly attractive to flies. Flies with CsChrimson (a red-shifted channelrhodopsin) expressed in these neurons will incessantly chase after red light . Importantly, their "interest to pursue" stimulation by red light is robust at single-animal level, does not attenuate over extended periods of time, and supersedes their interest in pursuing food and sex.
Second, using optogenetic activation of the presumed MFB-equivalent as "reward," we have found that Drosophila appear to be able to learn the causal relationship between their actions and the consequences. Using a closed-loop tracking and light-delivery system we developed (SkinnerTrax), we found that flies can learn a simple operant rule through trial and error so as to maximize receipts of stimulation of the presumed MFB-equivalent. Again, this learning is robust at single-animal level and develops quickly, thus allowing us to start probing its underlying genes and circuits. Our preliminary results suggest this form of learning and classical learning likely require different sets of genes and circuit components.
Third, we are developing a high-throughput apparatus for SkinnerTrax that allows "teaching" 40 individual flies simultaneously and we are gearing to produce at least 10 such apparatus so that we can examine learning of 400 individual flies per day. Armed with these apparatus, we will set out to pursue two goals. First, we will define the critical circuit components that enable this form of learning, taking advantage of the large collection of neuron-targeting tools available for Drosophila. Second, we will search for mutants -- by using both a forward genetics and a candidate-based approach -- that are outliers in learning. Specifically, we aim to find mutants whose learning capacity is significantly higher than that of WT flies.
Drosophila Decision-Making
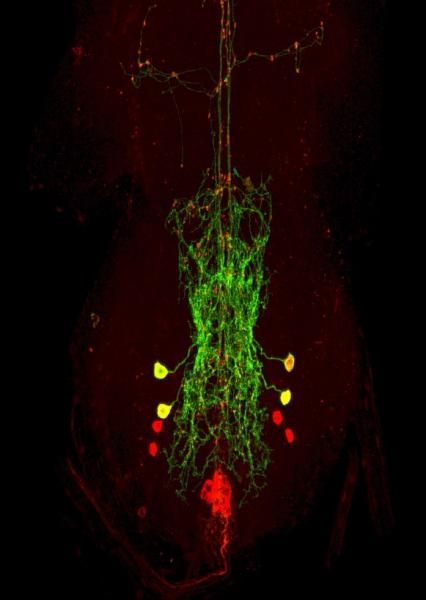
We are interested in understanding the circuit mechanisms by which animal brains decode the biological value (e.g., attractiveness) of sensory stimuli to guide simple decisions. We use Drosophila egg-laying site selection as our model system. We found that Drosophila females show clear preferences when tasked to rank the relative attractiveness of suitable egg-laying sites. To understand how the Drosophila brain assesses the relative attractiveness of sensory stimuli for egg-laying, we use a combined approach that includes high throughput behavioral screen using custom-built chambers, automated (machine vision) behavioral tracking of single animals (Movie 1), molecular genetic approaches to identify critical circuit components (Figure 1), and calcium imaging (Movie 2) and anatomical tracing techniques to determine the physical and functional connectivity of identified circuit components.
Yang Publications
Chen, Hsueh-Ling, Dorsa Motevalli, Ulrich Stern, and Chung-Hui Yang. “A functional division of Drosophila sweet taste neurons that is value-based and task-specific.” Proc Natl Acad Sci U S A 119, no. 3 (January 18, 2022). https://doi.org/10.1073/pnas.2110158119.
Imambocus, Bibi Nusreen, Fangmin Zhou, Andrey Formozov, Annika Wittich, Federico M. Tenedini, Chun Hu, Kathrin Sauter, et al. “A neuropeptidergic circuit gates selective escape behavior of Drosophila larvae.” Curr Biol 32, no. 1 (January 10, 2022): 149-163.e8. https://doi.org/10.1016/j.cub.2021.10.069.
Chen, Hsueh-Ling, Ulrich Stern, and Chung-Hui Yang. “Molecular control limiting sensitivity of sweet taste neurons in Drosophila.” Proc Natl Acad Sci U S A 116, no. 40 (October 1, 2019): 20158–68. https://doi.org/10.1073/pnas.1911583116.
Stern, Ulrich, Hemant Srivastava, Hsueh-Ling Chen, Farhan Mohammad, Adam Claridge-Chang, and Chung-Hui Yang. “Learning a Spatial Task by Trial and Error in Drosophila.” Curr Biol 29, no. 15 (August 5, 2019): 2517-2525.e5. https://doi.org/10.1016/j.cub.2019.06.045.
Wu, Shun-Fan, Ya-Long Ja, Yi-Jie Zhang, and Chung-Hui Yang. “Sweet neurons inhibit texture discrimination by signaling TMC-expressing mechanosensitive neurons in Drosophila.” Elife 8 (June 11, 2019). https://doi.org/10.7554/eLife.46165.
Kaneko, Takuya, Ann Marie Macara, Ruonan Li, Yujia Hu, Kenichi Iwasaki, Zane Dunnings, Ethan Firestone, et al. “Serotonergic Modulation Enables Pathway-Specific Plasticity in a Developing Sensory Circuit in Drosophila.” Neuron 95, no. 3 (August 2, 2017): 623-638.e4. https://doi.org/10.1016/j.neuron.2017.06.034.
Kaneko, Takuya, Ann Marie Macara, Ruonan Li, Yujia Hu, Kenichi Iwasaki, Zane Dunnings, Ethan Firestone, et al. “Serotonergic Modulation Enables Pathway-Specific Plasticity in a Developing Sensory Circuit in Drosophila.” Neuron 95, no. 3 (August 2, 2017): 722. https://doi.org/10.1016/j.neuron.2017.07.023.
Hu, Chun, Meike Petersen, Nina Hoyer, Bettina Spitzweck, Federico Tenedini, Denan Wang, Alisa Gruschka, et al. “Sensory integration and neuromodulatory feedback facilitate Drosophila mechanonociceptive behavior.” Nat Neurosci 20, no. 8 (August 2017): 1085–95. https://doi.org/10.1038/nn.4580.
Guntur, Ananya R., Bin Gou, Pengyu Gu, Ruo He, Ulrich Stern, Yang Xiang, and Chung-Hui Yang. “H2O2-Sensitive Isoforms of Drosophila melanogaster TRPA1 Act in Bitter-Sensing Gustatory Neurons to Promote Avoidance of UV During Egg-Laying.” Genetics 205, no. 2 (February 2017): 749–59. https://doi.org/10.1534/genetics.116.195172.
Gou, Bin, Edward Zhu, Ruo He, Ulrich Stern, and Chung-Hui Yang. “High Throughput Assay to Examine Egg-Laying Preferences of Individual Drosophila melanogaster.” J Vis Exp, no. 109 (March 24, 2016): e53716. https://doi.org/10.3791/53716.
Lab Members
Alumni
Yang Lab Open Positions
We are always happy to talk to outstanding candidates.
Contact Yang Lab
425 Bryan Research Bldg
Phone: 919-684-1494 (Rebecca), 919-684-1523 (lab)
Fax: 919-684-1738
Email: see People mailing address:
Bryan Research Bldg, Rm 425
311 Research Drive
Durham, NC 27710







