Kay Lab
We study how neural circuits devoted to specific visual processing tasks arise during development of the retina, and the consequences for circuit function when development goes wrong. The tools of mouse genetics are central to our approach, and we draw on a wide range of molecular, genetic, and imaging methods.
Wiring the retina and visual system -- from development to circuit function
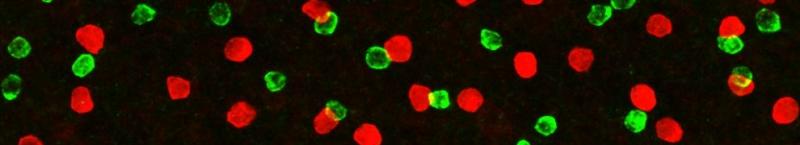
We study how the developing nervous system builds neural circuits. We seek to identify cellular and molecular mechanisms that generate different types of neurons, that endow them with specific identities, and that wire them together into circuits with a coherent function. Further, we would like to know what happens to circuit function when these mechanisms go awry, as may happen in neurodevelopmental disorders.
Here’s what motivates our research:
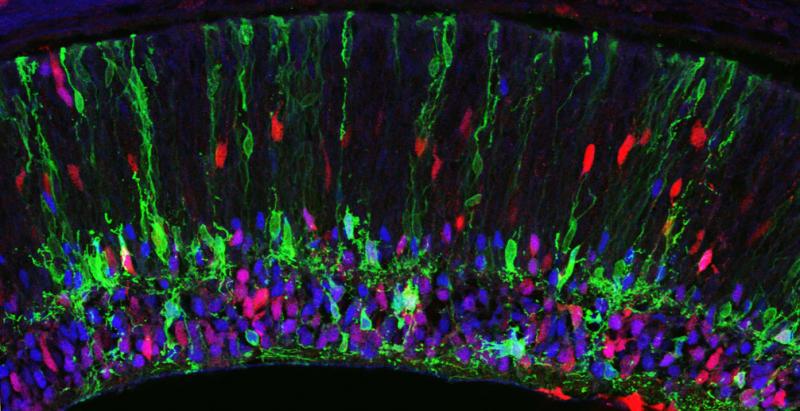
HOW DOES THE COMPLEX ANATOMY OF A NEURAL CIRCUIT RELATE TO ITS FUNCTION?
Many organs do their job just fine with a handful of cell types. But each individual part of the brain may have dozens of cell types – if you add that up for the whole central nervous system the number of cell types is easily in the thousands. Moreover, each of those cell types has its own unique dendrite and axon morphology, and unique patterns of connections with specific synaptic partners – this adds tremendously to the brain’s anatomical complexity. It makes intuitive sense, of course, that the brain needs complex anatomy to do all that it does. But we don’t yet understand the specifics – how do the particular anatomical features of particular circuits facilitate the encoding and transmission of information? This question is important not only for basic neuroscience, but also because there are developmental disorders that affect circuit anatomy and cognitive function – understanding why one leads to the other could help in devising treatments for these disorders.
 An ideal way to answer this question would be to study a model circuit that is easy to access and manipulate experimentally, that has a well-studied relationship between circuit structure and function, and whose synaptic connectivity is easy to measure anatomically and physiologically. That is why we study the mouse retina – it fits all these criteria, allowing us to use the tools of mouse genetics to subtly perturb circuit anatomy and make specific, testable hypotheses about the effects on circuit function. By taking advantage of all the retina has to offer as a model, we hope to pinpoint key developmental events and circuit wiring features that underlie not only visual function, but brain function more broadly.
An ideal way to answer this question would be to study a model circuit that is easy to access and manipulate experimentally, that has a well-studied relationship between circuit structure and function, and whose synaptic connectivity is easy to measure anatomically and physiologically. That is why we study the mouse retina – it fits all these criteria, allowing us to use the tools of mouse genetics to subtly perturb circuit anatomy and make specific, testable hypotheses about the effects on circuit function. By taking advantage of all the retina has to offer as a model, we hope to pinpoint key developmental events and circuit wiring features that underlie not only visual function, but brain function more broadly.
HOW DO GENES SET UP A BLUEPRINT FOR WIRING NEURAL CIRCUITS?
The wiring diagram of the retina, and indeed of the brain in general, is established during development by the coordinated maturation of each type of neuron within the tissue – each adopts a type-specific morphology and makes type-specific synaptic connections. Some of these maturation processes are driven by developmental programs intrinsic to each cell, while others involve highly specific cell-cell interactions that impart patterns of synaptic connectivity. If we can identify the genes required to implement these cell-intrinsic and collaborative events, we will move towards understanding at a fundamental level how circuits are assembled. Moreover, the molecules we identify may be candidate targets for therapeutic intervention in neurodevelopmental disorders.
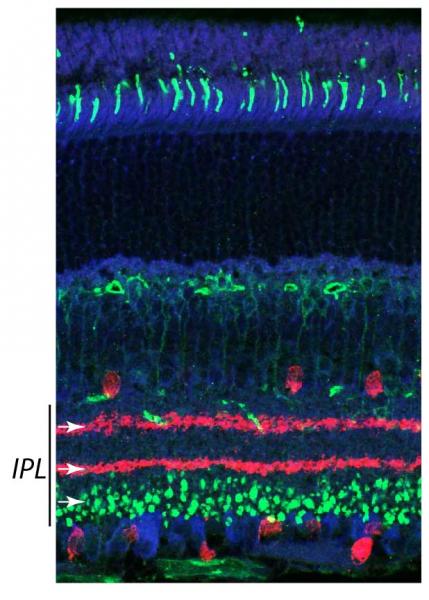
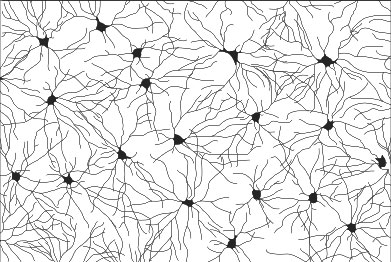 The retina is a convenient laboratory for discovering molecules that assemble circuits, because we can easily see details of circuit wiring that are hidden in other nervous structures. Retinal synapses are organized into parallel laminae – in the inner plexiform layer (IPL), where bipolar and amacrine interneurons synapse on retinal ganglion cells, there are as many as 10 separate sublayers (see figure at right/above). Most neurons project to a specific sublayer. Thus, a simple assay for synaptic partner choice is to ask whether a neuron alters its IPL stratification following a genetic experimental manipulation. Further, all retinal cell types show precise patterning in the plane of the visual field, which is required to prevent “blind spots” (see figure at left). By a simple screen for defects in axon, dendrite, or soma positioning across the retina, we can identify genes required for cell-type-specific neuronal interactions that assemble circuits. Moreover, because the mouse retina is uniquely amenable to genetic manipulation, we can conduct these screens in vivo – a major advantage of our system.
The retina is a convenient laboratory for discovering molecules that assemble circuits, because we can easily see details of circuit wiring that are hidden in other nervous structures. Retinal synapses are organized into parallel laminae – in the inner plexiform layer (IPL), where bipolar and amacrine interneurons synapse on retinal ganglion cells, there are as many as 10 separate sublayers (see figure at right/above). Most neurons project to a specific sublayer. Thus, a simple assay for synaptic partner choice is to ask whether a neuron alters its IPL stratification following a genetic experimental manipulation. Further, all retinal cell types show precise patterning in the plane of the visual field, which is required to prevent “blind spots” (see figure at left). By a simple screen for defects in axon, dendrite, or soma positioning across the retina, we can identify genes required for cell-type-specific neuronal interactions that assemble circuits. Moreover, because the mouse retina is uniquely amenable to genetic manipulation, we can conduct these screens in vivo – a major advantage of our system.
Kay Publications
Wang, Yuchen, Yan Cao, Cassandra L. Hays, Thibaut Laboute, Thomas A. Ray, Debbie Guerrero-Given, Abhimanyu S. Ahuja, et al. “Adhesion GPCR Latrophilin 3 regulates synaptic function of cone photoreceptors in a trans-synaptic manner.” Proceedings of the National Academy of Sciences of the United States of America 118, no. 45 (November 2021). https://doi.org/10.1073/pnas.2106694118.
Paisley, Caitlin E., and Jeremy N. Kay. “Seeing stars: Development and function of retinal astrocytes.” Dev Biol 478 (October 2021): 144–54. https://doi.org/10.1016/j.ydbio.2021.07.007.
Perelli, Robin M., Matthew L. O’Sullivan, Samantha Zarnick, and Jeremy N. Kay. “Environmental oxygen regulates astrocyte proliferation to guide angiogenesis during retinal development.” Development 148, no. 9 (May 1, 2021). https://doi.org/10.1242/dev.199418.
Ray, Thomas A., Kelly Cochran, Chris Kozlowski, Jingjing Wang, Graham Alexander, Martha A. Cady, William J. Spencer, et al. “Comprehensive identification of mRNA isoforms reveals the diversity of neural cell-surface molecules with roles in retinal development and disease.” Nat Commun 11, no. 1 (July 3, 2020): 3328. https://doi.org/10.1038/s41467-020-17009-7.
Luo, Lin, Mateusz C. Ambrozkiewicz, Fritz Benseler, Cui Chen, Emilie Dumontier, Susanne Falkner, Elisabetta Furlanis, et al. “Optimizing Nervous System-Specific Gene Targeting with Cre Driver Lines: Prevalence of Germline Recombination and Influencing Factors.” Neuron 106, no. 1 (April 8, 2020): 37-65.e5. https://doi.org/10.1016/j.neuron.2020.01.008.
Peng, Yi-Rong, Rebecca E. James, Wenjun Yan, Jeremy N. Kay, Alex L. Kolodkin, and Joshua R. Sanes. “Binary Fate Choice between Closely Related Interneuronal Types Is Determined by a Fezf1-Dependent Postmitotic Transcriptional Switch.” Neuron 105, no. 3 (February 5, 2020): 464-474.e6. https://doi.org/10.1016/j.neuron.2019.11.002.
Puñal, Vanessa M., Caitlin E. Paisley, Federica S. Brecha, Monica A. Lee, Robin M. Perelli, Jingjing Wang, Emily G. O’Koren, et al. “Large-scale death of retinal astrocytes during normal development is non-apoptotic and implemented by microglia.” Plos Biol 17, no. 10 (October 2019): e3000492. https://doi.org/10.1371/journal.pbio.3000492.
O’Koren, Emily G., Chen Yu, Mikael Klingeborn, Alicia Y. W. Wong, Cameron L. Prigge, Rose Mathew, Joan Kalnitsky, et al. “Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration.” Immunity 50, no. 3 (March 19, 2019): 723-737.e7. https://doi.org/10.1016/j.immuni.2019.02.007.
Ray, Thomas A., Kelly J. Cochran, and Jeremy N. Kay. “The Enigma of CRB1 and CRB1 Retinopathies.” Adv Exp Med Biol 1185 (2019): 251–55. https://doi.org/10.1007/978-3-030-27378-1_41.
Prigge, Cameron L., and Jeremy N. Kay. “Dendrite morphogenesis from birth to adulthood.” Curr Opin Neurobiol 53 (December 2018): 139–45. https://doi.org/10.1016/j.conb.2018.07.007.
Kay Lab Members
Alumni
Contact Kay Lab
Mailing address:
Duke University Eye Center
Box 3802
Durham, NC 27710
Shipping address:
Duke Eye Center
2351 Erwin Rd.
5th floor
Durham, NC 27705












